From Africa to Scotland
Sometime in our life each of us wonders, why am I here and where did I come from? Only you can define your life’s purpose and determine where you are going, but by analyzing the genes you inherited, I can tell you the amazing story of where you have been.
I can do this because each of us descended from a single man we call genetic Adam. Slight variations known as mutations occur at times as the Y chromosome is passed from father to son. Nucleotides are organic molecules that serve as the monomers, or subunits, of the nucleic acids DNA (deoxyribonucleic acid) and RNA (ribonucleic acid). The derived variants or mutations of nucleotides contained in the Y chromosome of human DNA is the science that allows us to compare and track male paternity.
There are tens of thousands of these nucleotides in the Y chromosome alone. Modern science has identified stable regions of the Y chromosome. From these stable regions we can now detect mutations at each stable nucleotide. Since a single nucleotide can be identified, we call these variations single nucleotide polymorphisms or SNP.
We use four of the subunits, adenine, cytosine, guanine and thymine [A-C-G-T] to determine when there is a variation or mutation from an ancestral position to a derived position. For example, the ancestral position may be A and the derived position is T. Each of these positions is located within a stable region and is accompanied by a starting point and an end point. The derived position is identified by a letter and number. This identification is always started with the haplogroup letter followed by the testing lab that identified the mutation followed by an identification number.
As example I employ one of my SNP’s, R-Y28576. R = I am of haplogroup R. Y = Y Full, the lab that identified the derived mutation. 28576 = the identification number issued by Y Full. The actual position of this SNP is 7783526 and is found in region 7783475-7783576. The ancestral position is G and the derived mutation is T. This SNP is also known as YFS2081410.
The forward and reverse primer for R-Y28576 is Z2564_F and Z2564_R. The comments for this SNP are downstream of FGC17354 and peer to FGC17347. This is an R1b category of U106 subclade with the following phylogeny Z7 > FGC7559 > FGC17354 > Y28576.
This may all seem entirely too complex for many. For those of us trained to understand, it’s like GPS leading to a location on the tree of humanity. Don’t sweat the details; I’m just explaining how I am able to tell you where you have been.
Genetic Adam
For our purposes genetic Adam was a man of haplogroup A00, born about 200,000 years ago in the Rift Valley of Africa, during the middle Paleolithic. The oldest fossilized remains were found in modern Ethiopia and dated to 195,000 years ago. Adam was much like his hominin neighbors and probably existed in very much the same way. He is the man we recognize as the founding event of Homo sapiens. His increased cranial capacity would however, give him a tremendous advantage over his competitors.
His capacity for abstract thought was greater. Though he probably did not invent the technology available to him and his contemporaries, he would be able to make improvements and adjustments faster and better. He probably developed superior strategies for hunting, preserving and tanning. Improved weapons and clothing would give advantage concerning environment and protection. The advancement of technology and strategy may have played a role in his rapid proliferation as he was able to support and sustain larger tribal units.
It may also be the reason we are writing about his Y DNA and his descendants instead of Homo erectus, Neanderthal or the Denisnovans. Somehow Adam was the survivor and we see traces of other hominins in our modern admixture. The survival of hominin DNA in modern humans suggests all hominins were of the same species and not merely the same genus. Cross genus hybrids are sterile and can’t procreate. There would be no traces of ancient hominin DNA in modern humans if this was the case.
African or Non-African
This is a confusing term in Y DNA studies since all modern Y DNA originated in Africa. As mentioned earlier, polymorphisms occurred in African populations from A00 genetic Adam. Since we are now so far removed from the founding event the most basal subclades of A are rare. Over a span of about 60,000 years the African haplogroups took shape. The chart below accurately defines African and non African haplogroups.
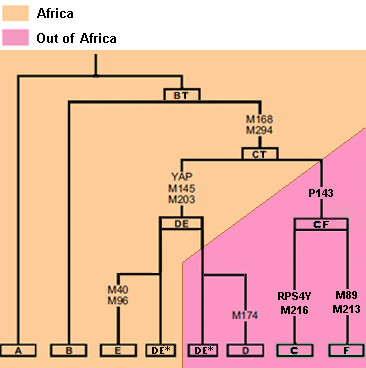
For our purpose the phylogeny is A0-T > A1 > A1b > BT > CT > CF > F.
BT, CT and CF are now fossil haplogroups. We know they existed because men test positive at the SNP’s contained within their defining boundaries. This is accomplished by comparison of SNP’s shared by the downstream haplogroups. For example, C and F continue to mutate and are living haplogroups. CF is the combination of SNP’s possessed by both C and F men.
We can conclude haplogroup CF was the parent of European ancestor F. The exact geographic origin of CF is unknown, but it was almost certainly in the fertile crescent of North Africa or Southwest Asia. Age estimate of emergence for CF is 71,800 - 62,500ybp - about the time modern humans were coming out of Africa.
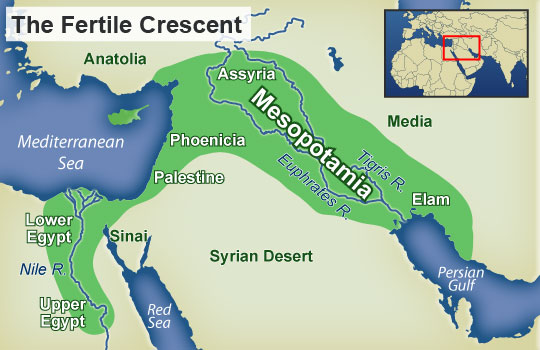
The Near Extinction of Man
NASA evidence has indicated there was once a mega lake in the Eastern Sahara desert that started forming about 250,000 years ago. This was during a cycle of abundant rain in the regions ancient past. At its peak it covered about 42,000 square miles. About 100,000 years ago it may have opened up a viable conduit for human migration to the Mediterranean. The next cycle of abundant rain would be between 14,000 and 6,000 years ago.
As the water slowly dried up and the Nile shifted back into its familiar banks, people were also forced to retreat into the narrow Nile Valley corridor.
Decreasing habitat, disease and the eruption of super volcano Toba in Indonesia about 74,000 years ago may have combined to produce the near extinction event. This could have spurred human migration out of Africa through the Nile Valley into Eurasia.
The hunter gatherer lifestyle usually could not support more than 30 people per square mile. It was a nomadic lifestyle that required constant movement as game, fish and resources diminished in an area. During this period populations of North Africa and the Nile Valley decreased to just a few people per square mile. Migration out of North Africa at this time may have been a necessity for survival.
The separation of African and Non African Y DNA was complete. African DNA was cut off and isolated in sub-Sahara Africa. With abundant water and resources there was no reason to venture into a desert region larger than the continental United States.
The Super Cluster
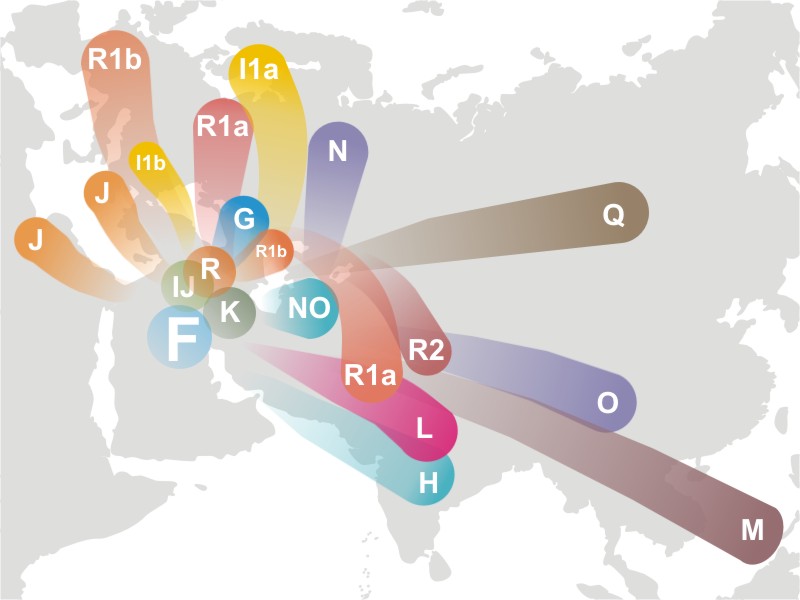
Haplogroup F is a living haplogroup found all over Europe today. Its defining mutations are F-M89, P14 and M213. As indicated in the illustration above, haplogroup F is the parent of several European haplogroups. The super cluster of GHIJK was the direct descendant of F. GHIJK formed between 51,400 - 46,300 years ago. GHIJK does not exist today but is defined by SNP’s common to G, H, I, J and K. All men of these haplogroups are also positive for M89. Approximately 90% of men today are descendant of haplogroup F
Haplogroup G was the first to diverge. Its defining mutation is G-M201. Emergence of G 50,900 - 46,200 years ago. The divergence from the super cluster is estimated from Northern Iran to Georgia and the caucus region.
Haplogroup H was the second to diverge from patent HIJK. Its defining mutation is H - L901 / M2939. Emergence of H is in the same era as G, 50,900 - 46,200 years ago. H1 M69 is rarely found outside the Indian sub continent. H2 P96, L279, M282 is the European subclade. H3 Z5857 is found in South Asians.
The Diversification of Haplogroup K
Haplogroup K is the next to diverge. Its defining mutation is K - M9. Basal haplogroup K is rare in modern times and found in low frequencies in Eurasia, Oceania and Africa. The Emergence of K M9 was 49,700 - 44,600 years ago. Haplogroup K descendants are the most diverse by far. Its descendants are also found on every habitable continent of the world.
The descendant haplogroups of K came rapidly as K has only 17 equivalent mutations.
Direct descendant LT [aka K1] is defined by mutation L298/P326. Emergence of LT L298 is 49,600 - 41,400 years ago. LT is no longer a living haplogroup but is survived by haplogroups L and T.
Haplogroup L is defined by mutation L - M20. Emergence of L M295 is 45,500 - 39,700 years ago. L is predominantly Middle Eastern with high concentrations in Pakistan. L1 M27 is found in South Asia. L2 M317 is found in low frequencies in Central Asia, Southwest Asia and Southern Europe. L2 - L595 is extremely rare and is seldom found outside of Western Europe.
Haplogroup T is defined by mutation T - M184. Emergence of T M184 is the same as L, 45,500 - 39,700 years ago. Basal T1 is found primarily in the Eastern Mediterranean basin. T2 PH110 is found in Northern European countries, Georgia and the Kingdom of Bhutan east of Nepal.
Haplogroup K2 is another direct descendant of haplogroup K and is defined by mutation K - M526. Emergence of K M526 is 49,600 - 44,600 years ago. Basal K M526 is found today in aboriginal Australians. K4 and C4 are the dominant haplogroups of Aborigines. Incidentally, both are descendants of haplogroup CF.
Haplogroup NO is descent from K2 and is defined by mutation NO - M214. It emerges rapidly after K2 and is the parent of haplogroups N and O. The divergence period of N and O is rapid since NO has only 17 mutation equivalents. NO is no longer a living haplogroup.
Haplogroup N is defined by mutation N - M231. Emergence of N M231 is 39,300 - 34,900 years ago. N3 M46 is the haplogroup found in modern populations of northern Eurasia.
Haplogroup O is defined by mutation O - M175. Emergence of O M175 is the same era as N, 39,000 - 34,900 years ago. Modern populations range from north central Asia to highest concentrations in Southeast and Eastern Asia.
Haplogroup K2b is perhaps the shortest lived haplogroup on earth with only 2 equivalent mutations and only about 3,000 years younger than K - M49. The defining mutation is K2b - P331. Since this is no longer a living haplogroup it is best known as MPS representing its three descendant haplogroups.
K2b1 descendants
Haplogroup M is defined by mutation M - P256. Emergence of M P256 is relative to its sibling clades, 49,600 - 41,400 years ago. Haplogroup M is found primarily in eastern Indonesia or New Guinea.
Haplogroup S is defined by mutation S - M230. Its emergence and distribution are synonymous with haplogroup M with higher concentrations in New Guinea.
Haplogroup P is the European ancestor and was formerly known as K2b2. The defining mutation is P - P295. Once thought dead, a group positive at P - P109 has emerged and currently has 420 members. Its emergence is so near K2b1 descendants it is not worth mentioning. P is best known for its descendants, haplogroups Q and R.
Haplogroup Q is defined by mutation Q - M242. Emergence of Q M242 is 34,000 - 29,800 years ago. It has one major subclade, Q1 - L232 and is found in very low frequencies in Europe. It increase across Siberia and becomes dominant for Native American people. Highest frequencies occur in the Western United States and Canada and all across South America.
Haplogroup R is defined by mutation R - M207. Its emergence was in the same era as Q. It is so universal it needs its own map.
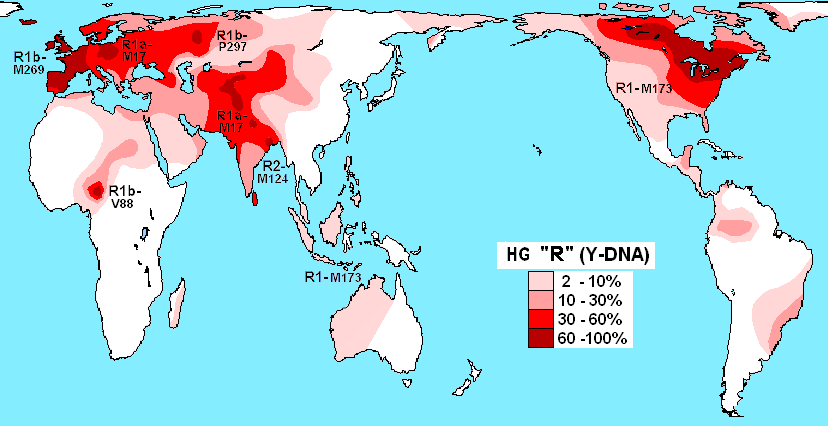
R1b - M269 dominates Western Europe. R1a has high concentrations in Russia, Eastern and Central Europe and Scandinavia. As you can see R1 - M173 is found in Native American populations of Eastern Canada and the United States - this is the root of R1. R1a - M17 has high concentrations in Central Asia. We will not delve into the hundreds of subclades of each category of R in this study. We tend to find R and Q in the same populations to varying degrees.
Haplogroup IJ
I didn’t forget this last group to emerge from the super cluster and its descendants I and J. IJ is defined by mutation IJ - M429/P125. IJ emerged in the same era as its only sibling K, 49,600 - 44,600 years ago.
Haplogroup I is defined by mutation I - M170. I M170 emerged 45,700 - 43,300 years ago. As the distribution map above indicates emergence could have taken place from Eastern Turkey / Northern Iraq to the Balkans in Europe. Basal haplogroup I is rare comprising less than 2% of European populations. Subclades I1 and I2 are more prevalent. I was the first out of Africa haplogroup to reach Europe in significant numbers. There is debate concerning whether it emerged in Europe or West Asia.
Haplogroup J is defined by mutation J - M304. J M304 emerged 45, 700 - 40,200 years ago in Western Asia. Though J is primarily a Middle Eastern haplogroup, subclades are distributed around the Mediterranean. Basal J is extremely rare today. J2a1b - M67 is found in low percentages in southern Europe from the Balkans to the British Isles.
Haplogroup DE
Our Scottish members of haplogroup E are the only ones not included in the super cluster.
Haplogroup DE most likely formed in Africa. As you can see from our chart of African / Non African, a split occurred. This resulted in different mutations of haplogroup E descendant of DE. E1a is almost exclusively Sub - Saharan and considered African. E1b is almost exclusively North African / European.
Since our focus is on subclades that reached Scotland, we fast forward to E - V13 defined by mutation L542. E - V13 is the most abundant subclade of haplogroup E to reach the main Isle and Scotland. E V13 L452 emerged between 8900 - 6400 years ago.
Many folks, including participants of our Forrest / Forrester project, are listed as E - L117. This position is also known as E - M35 and is the genetic grandparent of E - V13. This places E - L117 as a clade head with multiple mutations of descent. E - L117 as lowest tested SNP requires additional SNP testing to determine their haplogroup.
Probable but not carved in Stone
As we explore probable times and places of emergence, keep in mind further evidence could, and probably will, change our current understanding. The hypothesis for each group covered is based on the best information we have available today - it could change tomorrow.
Please remember my stance is everything I just wrote of is probably wrong. There are simply not enough ancient DNA samples available for the work to be exhaustive or conclusive. With that in mind, please enjoy the probable migrations of the various haplogroups from Africa to Scotland.
Nathan Forrister
Clan Forrester Society
DNA Research Coordinator